Definition of Mitochondrial DNA
Miscellanea / / July 04, 2022
concept definition
The mitochondrial genome is a small, closed, double-helix circular molecule of 16,569 base pairs (bp) that is inherited only from mothers to sons and daughters (rarely). cases of heteroplasmy), so there is no genetic recombination at the time of conception, it also evolves exclusively by accumulation of mutations in the weather.






Lic. in Physical Anthropology
The high rate of mutation of the mitochondrial genome (10 to 20 times that of the DNA nuclear, in terms of genes with comparable functions) is useful for differentiating between populations that over time have been biologically related, however, their mutation rate can be so fast that mutation phenomena can occur. backmutation. The rate of evolution The mean of this molecule has been calculated from species in which divergence times from fossil remains were available, and from biodemographic or protein data; the result gives 1-2% per million years, valid for different orders.
This particular DNA functions as a tool to research the biological kinship between populations because their properties allow elucidating the relationships between populations that have diverged in recent times, without having to consider recombination phenomena and adding a dimension temporary; For this, the following must be kept in mind: two individuals whose common ancestor has been a woman will have mitochondrial DNA molecules as different as time has elapsed since the separation of the ancestor.
Haplogroups for mitochondrial DNA classification in geographic distribution
Some regions of DNA can be very similar to each other, allowing their classification in the same cluster, which is known as a haplogroup. For example, Torroni et al (1993) typified the founding mitochondrial lineages for the American Continent, the which received the denomination of haplogroups A, B, C, and D, according to point mutations of the sequence mitochondrial. These mutations create different cleavage sites for specific enzymes, as described below: in lineage A there is the site cutoff 663 for the enzyme HaeIII, lineage C is characterized by site 13259 for HincII, in lineage D site 5176 is recognized for AluI; this identification is made from fragment length polymorphisms (RFLP). In the case of lineage B, there is a deletion of 9 base pairs at position 8272-8289.
The geographical distribution of each lineage has been described as follows: lineage A is predominant in the American Continent and especially in North America; however, this haplogroup is also attributed to Mesoamerican populations. Lineages C and D appear mainly in South America; lineage B is found in the northern and southern region of the Pacific coast, and Kemp et al (2010) proposes it as a characteristic lineage of the family Yuto-Aztecan and southwestern U.S.A.
In addition, there are mutations that are shared between individuals of the same haplogroup, which has made it possible to describe specific haplotypes (or sublineages) of some populations. Figure 1 shows the regions of the mitochondrial DNA that contain the mutations to recognize each lineage.
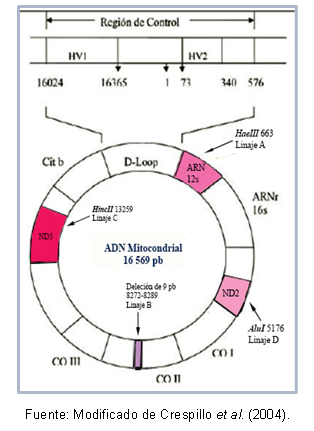
Figure 1. Mitochondrial DNA regions typifying each American founding lineage.
For some years only these four lineages were recognized until haplogroup X was included for the northern populations of the continent; which is also distantly related to European populations. The previous lineages have a correspondence with Asian populations, although they are observed less frequently than in the American Continent; lineages A, B, and C are not found in modern Africans and Caucasians; and lineage D also exists in Africa, but associated with other restriction sites. With the above, we could say that these haplogroups characterize the American populations and therefore their study is appropriate in terms of settlement and migrations (old and recent).
genetic distances
The way to establish genetic similarities or differences between populations is with genetic distances, which can have a historical explanation, since they change (increase or decrease) over the generations, and can lead us to events in history of one population for example: at the time of a great migration or contact between two cultures, with this calculation we can discern what mechanisms came into action to give us this or that result.
Both genetic drift and gene flow have to do with the ease with which a group, in this case human beings, has to move within a group. territory and to come into contact with other groups. So, geographic isolation is the genetic distance that grows with increasing geographic distance between human groups.
Mitochondrial DNA diversity hypothesis
To explain the diversity in the founding mitochondrial lineages in America, there are two hypothesis: that this continent has been colonized by multiple events from Beringia, or that once migration has occurred, evolutionary changes have occurred after colonization. Two routes of entry of these variations to the continent are also explained, the first proposes that the four founding haplogroups without variations, that is, each one with a root haplotype, they could have arrived just after the Last Glacial Maximum or a little before, with dates of 21 thousand to 19 thousand years ago and would have followed a coastal route by the Peaceful; the second proposal suggests that these intra-haplogroup variations already existed in Beringia and were brought to the south of the American continent but their entry would have been exactly at the end of the Last Glacial Maximum so that the routes within the continent would already be free, then the entry of these human groups would be 19 thousand ago years. There is also a great diversity of haplogroup A and a coalescence time for it that is shorter than the rest (17 thousand years).
The most likely explanation for this is that it is due to secondary expansions of haplogroup A from Beringia, long after the end of the Last Glacial Maximum. Despite the discrepancy regarding the time of man's entry into the Americas, genetic studies have give some clarity, because they support the hypothesis that there were human groups in the American continent before Clovis; and it is found that there is a separation between the ancestors of Northeast Asia 25-35 thousand years ago and the entry into America 15-35 thousand years ago. Figure 2 shows the routes of the mitochondrial haplogroups in the world and the time of divergence in years before the present.
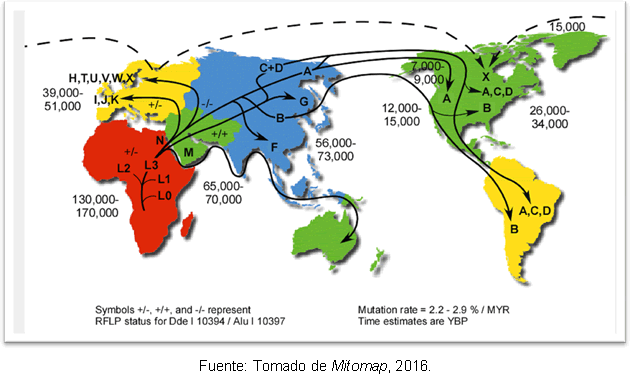
Figure 2. Map of the different routes of dispersal of mitochondrial lineages
Importance of phylogeography in genetic study and mitochondrial gene data
There is a very useful tool for genetic analysis; the phylogeography. These are the first applications of molecular studies, and with it we seek to be able to determine the phylogenetic and spatial relationships between nucleotide sequences, in this case DNA mitochondrial. The spatial distribution may resemble a temporal pattern, that is, the most geographically distant DNA sequences should be the most genetically different, as well as DNA sequences that diverged long ago should also be the most different genetically. So geographically distant populations, with little or no gene flow between them, would accumulate differences due to genetic drift and mutation, even by selection; but situations may occur that do not allow drift, such as one or more founder effects or other patterns of gene flow.
The data provided by mitochondrial genes has been one of the most useful in phylogeography research due to the characteristic that these genes have of not recombining and as a result they show a much clearer phylogenetic line than many nuclear genes. The effective population size calculated with the mitochondrial genes (and with the Y chromosome functioning in a similar way) is about a quarter of that calculated for the mitochondrial genes. nuclear genes, then divergence occurs almost four times faster than with nuclear genes, this rapid rate of divergence (and gene flow) can cause the observed pattern of inheritance in these uniparental genes is different than the phylogenies obtained with nuclear genes (which represent the majority of the gene pool in a individual).
References
Achilli, A., Perego, U. A., Bravi, C. M., Coble, M. D., Kong, Q.-P., & et al. (2008). The Phylogeny of the four Pan-American MtDNA Haplogroups: Implications for Evolutionary and Disease Studies. PLoS ONE, 3(3), e1764.Cavalli-Sforza, L. L. (2000). Genes, peoples and languages (1st Edition in Pocket Library ed.). Barcelona, Spain: Critical Editorial.
Rebato, E., Susanne, C., & Chiarelli, B. (Edits.). (2005). To understand Biological Anthropology: evolution and human biology. Divine Word.
Torroni, A., Schurr, T. G., Cabell, M. F, Brown, M. D, Neel, J. V., Larsen, M., et al. (1993). Asian affinities and continental radiation of the four founding Native American mtDNAs. The American Journal of Human Genetics, 53(3), 563.
(2022). mitochondrial DNA, National Human Genome Research Institute.
-Mitochondrial DNA schematic reference-
Crespillo, M., Paredes, M., Arimany, J., Guerrero, L., & Valverde, J. (2004). Spanish Civil War (1936-1939): identification of human remains from mass graves in Catalonia by mitochondrial DNA analysis. About a case. Forensic Medicine Notebooks (38), 37-46.



